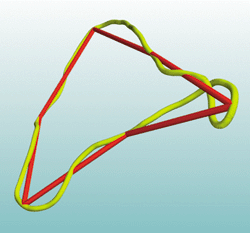
Fig. 1. DNA model.
Piece-wise linear models of polymers were introduced by Kuhn in the 1930s to provide a tractable model simpler than the continuous description that he knew was closer to reality. His approximation and subsequent refinements have successfully allowed relatively simple mathematical and numerical treatments of physical phenomena in DNA. However, the effective physical properties of DNA are relatively uniform along the backbone and, in reality, there are no hinges interspersed with rigid regions (unless perhaps one reduces to a single base pair-level description).
Electron microscopy of DNA has shown that on the wide range of viewable length scales the double helix appears as a continuous curve (Fig. 1). Any such smooth curve (in yellow) can be well approximated by n line segments, with n sufficiently large (the case n = 6 shown in red seems inadequate). The question of how large n needs to be depends on the smoothness of the underlying curve, or whether the spaghetti is cooked al dente or scotti. For models of DNA, this smoothness depends on the number of base pairs represented by the continuous curve (2686 for the data shown).

Fig. 1. DNA model.
We know of no realistic application involving a piece-wise linear model of a DNA loop where the number of links, n, can be as few as 6. Moreover, any conclusion that depends sensitively on n--for example, rigidity--for n = 6, which disappears for n = 7, cannot be physically pertinent for DNA.
Andrzej Stasiak
Jacques Dubochet
Université de Lausanne,
CH-1015
Lausanne, Switzerland.
E-mail:
Andrzej.Stasiak@lau.unil.ch;
Jacques.Dubochet@lau.unil.ch
Patrick Furrer
Oscar Gonzalez
John Maddocks
Ecole Polytechnique Federale de Lausanne,
CH-1015 Lausanne, Switzerland.
E-mail:
pfurrer@masg1.epfl.ch;
gonzalez@dma.epfl.ch;
maddocks@dma.epfl.ch
The point of Stasiak et al. that DNA does not resemble a polygon of 6 or 7 rigid segments is absolutely correct. Such a model may be more realistic for an artificially synthesized polymer, and several mathematicians I spoke with suggested that this might be an interesting project for chemists to work on (similar to the synthesis of "rotaxanes," in which two loops are geometrically linked without being topologically linked). For DNA, the best model may indeed be one that depends on the details of how the spaghetti is cooked, as Stasiak et al. suggest.
The mathematical treatment of large molecules with a certain amount of rigidity is still very much in its infancy. If mathematicians continue to study models of molecules that are in some respects outdated or oversimplified, the reason is that there are still interesting questions about them that have not been answered. The mathematicians I spoke with felt that the shape presented by Jason Cantarella was a useful step toward understanding the effect of rigidity on conformation. The conclusion: Rigidity matters. --Dana Mackenzie